As well as working with local in-memory data stored in data frames, dplyr also works with remote on-disk data stored in databases. This is particularly useful in two scenarios:
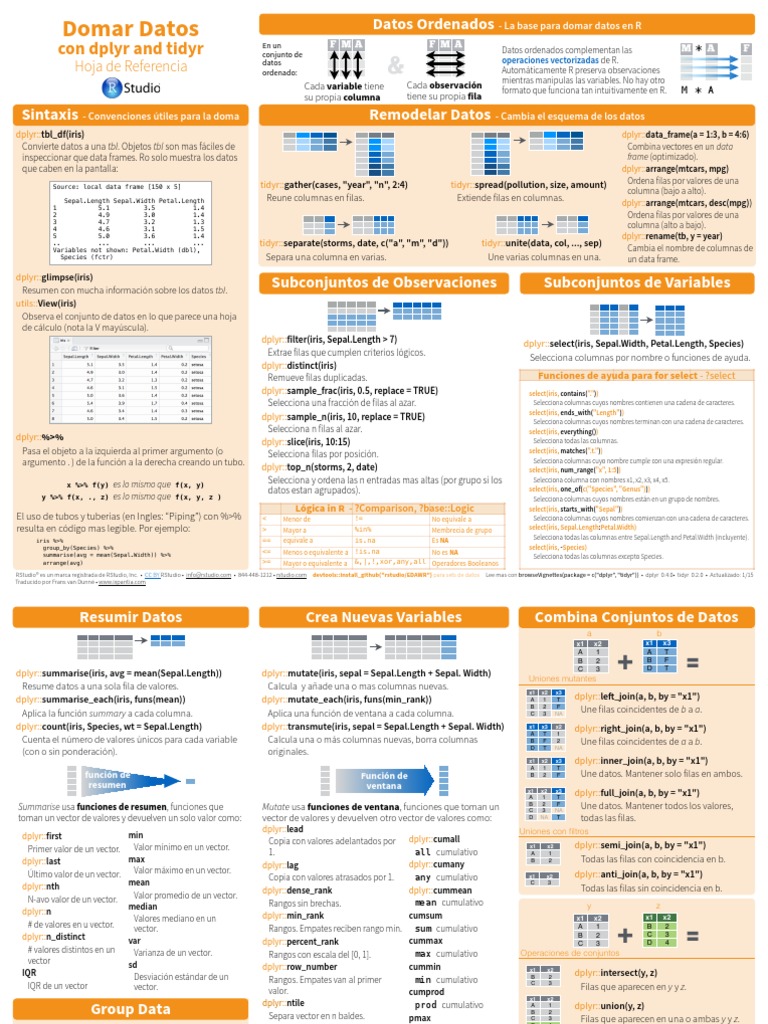
Dplyr‘s groupby function can group together rows of a data frame with the same value(s) in either a specified column or multiple columns, allowing for the application of summary functions on the individual groups. Groupby changes the unit of analysis from a complete dataset to individual groups. For example, consider a data frame countries. There are lots of Venn diagrams re: SQL joins on the internet, but I wanted R examples. Those diagrams also utterly fail to show what’s really going on vis-a-vis rows AND columns. Other great places to read about joins: The dplyr vignette on Two-table verbs. The Relational data chapter in R for Data Science (Wickham and Grolemund 2016.
Your data is already in a database.
You have so much data that it does not all fit into memory simultaneouslyand you need to use some external storage engine.
(If your data fits in memory, there is no advantage to putting it in a database; it will only be slower and more frustrating.)
This vignette focuses on the first scenario because it is the most common. If you are using R to do data analysis inside a company, most of the data you need probably already lives in a database (it’s just a matter of figuring out which one!). However, you will learn how to load data in to a local database in order to demonstrate dplyr’s database tools. At the end, I’ll also give you a few pointers if you do need to set up your own database.
Getting started
To use databases with dplyr, you need to first install dbplyr:
You’ll also need to install a DBI backend package. The DBI package provides a common interface that allows dplyr to work with many different databases using the same code. DBI is automatically installed with dbplyr, but you need to install a specific backend for the database that you want to connect to.
Five commonly used backends are:
RMySQLconnects to MySQL and MariaDB
RPostgreSQLconnects to Postgres and Redshift.
RSQLite embeds a SQLite database.
odbc connects to many commercialdatabases via the open database connectivity protocol.
bigrquery connects to Google’sBigQuery.
If the database you need to connect to is not listed here, you’ll need to do some investigation yourself.
In this vignette, we’re going to use the RSQLite backend, which is automatically installed when you install dbplyr. SQLite is a great way to get started with databases because it’s completely embedded inside an R package. Unlike most other systems, you don’t need to set up a separate database server. SQLite is great for demos, but is surprisingly powerful, and with a little practice you can use it to easily work with many gigabytes of data.
Connecting to the database
To work with a database in dplyr, you must first connect to it, using DBI::dbConnect(). We’re not going to go into the details of the DBI package here, but it’s the foundation upon which dbplyr is built. You’ll need to learn more about if you need to do things to the database that are beyond the scope of dplyr.
The arguments to DBI::dbConnect() vary from database to database, but the first argument is always the database backend. It’s RSQLite::SQLite() for RSQLite, RMySQL::MySQL() for RMySQL, RPostgreSQL::PostgreSQL() for RPostgreSQL, odbc::odbc() for odbc, and bigrquery::bigquery() for BigQuery. SQLite only needs one other argument: the path to the database. Here we use the special string, ':memory:', which causes SQLite to make a temporary in-memory database.
Most existing databases don’t live in a file, but instead live on another server. In real life that your code will look more like this:
(If you’re not using RStudio, you’ll need some other way to securely retrieve your password. You should never record it in your analysis scripts or type it into the console.)
Our temporary database has no data in it, so we’ll start by copying over nycflights13::flights using the convenient copy_to() function. This is a quick and dirty way of getting data into a database and is useful primarily for demos and other small jobs.
As you can see, the copy_to() operation has an additional argument that allows you to supply indexes for the table. Here we set up indexes that will allow us to quickly process the data by day, carrier, plane, and destination. Creating the write indices is key to good database performance, but is unfortunately beyond the scope of this article.
Now that we’ve copied the data, we can use tbl() to take a reference to it:
When you print it out, you’ll notice that it mostly looks like a regular tibble:
The main difference is that you can see that it’s a remote source in a SQLite database.
Generating queries
To interact with a database you usually use SQL, the Structured Query Language. SQL is over 40 years old, and is used by pretty much every database in existence. The goal of dbplyr is to automatically generate SQL for you so that you’re not forced to use it. However, SQL is a very large language, and dbplyr doesn’t do everything. It focuses on SELECT statements, the SQL you write most often as an analyst.
Most of the time you don’t need to know anything about SQL, and you can continue to use the dplyr verbs that you’re already familiar with:
However, in the long run, I highly recommend you at least learn the basics of SQL. It’s a valuable skill for any data scientist, and it will help you debug problems if you run into problems with dplyr’s automatic translation. If you’re completely new to SQL, you might start with this codeacademy tutorial. If you have some familiarity with SQL and you’d like to learn more, I found how indexes work in SQLite and 10 easy steps to a complete understanding of SQL to be particularly helpful.
The most important difference between ordinary data frames and remote database queries is that your R code is translated into SQL and executed in the database, not in R. When working with databases, dplyr tries to be as lazy as possible:
It never pulls data into R unless you explicitly ask for it.
It delays doing any work until the last possible moment: it collects togethereverything you want to do and then sends it to the database in one step.
For example, take the following code:
Surprisingly, this sequence of operations never touches the database. It’s not until you ask for the data (e.g., by printing tailnum_delay) that dplyr generates the SQL and requests the results from the database. Even then it tries to do as little work as possible and only pulls down a few rows.
Behind the scenes, dplyr is translating your R code into SQL. You can see the SQL it’s generating with show_query():
If you’re familiar with SQL, this probably isn’t exactly what you’d write by hand, but it does the job. You can learn more about the SQL translation in vignette('sql-translation').
Typically, you’ll iterate a few times before you figure out what data you need from the database. Once you’ve figured it out, use collect() to pull all the data down into a local tibble:
collect() requires that database does some work, so it may take a long time to complete. Otherwise, dplyr tries to prevent you from accidentally performing expensive query operations:
Because there’s generally no way to determine how many rows a query willreturn unless you actually run it,
nrow()is alwaysNA.Because you can’t find the last few rows without executing the wholequery, you can’t use
tail().
You can also ask the database how it plans to execute the query with explain(). The output is database-dependent and can be esoteric, but learning a bit about it can be very useful because it helps you understand if the database can execute the query efficiently, or if you need to create new indices.
Creating your own database
If you don’t already have a database, here’s some advice from my experiences setting up and running all of them. SQLite is by far the easiest to get started with, but the lack of window functions makes it limited for data analysis. PostgreSQL is not too much harder to use and has a wide range of built-in functions. In my opinion, you shouldn’t bother with MySQL/MariaDB; it’s a pain to set up, the documentation is sub par, and it’s less feature-rich than Postgres. Google BigQuery might be a good fit if you have very large data, or if you’re willing to pay (a small amount of) money to someone who’ll look after your database.
All of these databases follow a client-server model - a computer that connects to the database and the computer that is running the database (the two may be one and the same, but usually aren’t). Getting one of these databases up and running is beyond the scope of this article, but there are plenty of tutorials available on the web.
MySQL/MariaDB
In terms of functionality, MySQL lies somewhere between SQLite and PostgreSQL. It provides a wider range of built-in functions, but it does not support window functions (so you can’t do grouped mutates and filters).
PostgreSQL

PostgreSQL is a considerably more powerful database than SQLite. It has:
a much wider range of built-in functions, and
support for window functions, which allow grouped subset and mutates to work.
BigQuery
BigQuery is a hosted database server provided by Google. To connect, you need to provide your project, dataset and optionally a project for billing (if billing for project isn’t enabled).
It provides a similar set of functions to Postgres and is designed specifically for analytic workflows. Because it’s a hosted solution, there’s no setup involved, but if you have a lot of data, getting it to Google can be an ordeal (especially because upload support from R is not great currently). (If you have lots of data, you can ship hard drives!)
4 min read2020/04/16Motivation
I use R to access data held in Microsoft SQL Server databases on a daily basis. As a result of running into problems, I’ve realized I don’t have an understanding of the roles different components, notably dplyr, dbplyr, odbc and dbi each play in the process. This contributes to the fact that my efforts to resolve or mitigate issues are often an inefficient combination of Google searches and trial and error. Additionally, as I find or develop workarounds, I am unwilling to promote them with others because I don’t fully understand the cause of the issue and, as a result, I am not confident I have addressed the problem at the appropriate level.
Specifically, this is most motivated by the, “Invalid descriptor index,” error documented here.
I am writing what I learn to solidly my thinking and to help others who may experience the same challenge.
Scope
Given that the source of my motivation is encountering problems when working with data in Microsoft SQL Server databases, and that I prefer to use packages in the tidyverse, this investigation will be focused on how these packages work together to collect data from databases managed by Microsoft SQL server.
I won’t be writing about other options like RODBC, RJDBC or database-specific packages.
I will also not include comments about database management systems (DBMS) other than Microsoft SQL Server.
While it’s possible to generalize many of the concepts I write about here to other DBMS systems, I will not explicitly call them out. There are plenty of resources that do that. I aim to be very focused on how these components interact in a tidyverse and Microsoft SQL Server environment in the hopes it will help paint a simpler, clearer picture for others working in that same configuration.
Additionally, I almost never write data to a DBMS, and I suspect this is the case for many people working as analysts in Enterprise environments. In light of that I will be focused on how these components work together to extract data from SQL, and not how they write data to it.
Role of dplyr and dbplyr packages
dbplyr is the database back-end for dplyr - it does not need to be loaded explicitly, it is loaded by dplyr when working with data in a database.
dbplyr translates dplyr syntax into Microsoft SQL Server specific SQL code so dplyr can be used to retrieve data from a database system without the need to write SQL code.
dbplyr relies on the DBI and odbc packages as an intermediaries for connections with a SQL Server database.
Tidyverse Cheat Sheet Pdf
Dplyr can also pass on explicitly written (not translated from dplyr) SQL code to DBI.
dbplyr generates, or captures, the SQL code that is then passed into the front-end of the database stack provided by DBI, odbc and the ODBC driver
Role of the DBI package
DBI segments the connectivity to the SQL database into a, “front-end,” and a, “back-end.”
DBI implements a standardized front-end to dbplyr, and the odbc package acts as a driver for DBI to interface with SQL Server.
An example of front-end functionality provided by DBI…
- connect/disconnect to the database
- create and execute statements in the DBMS
- extract results/output from statements
- error/exception handling
- information (meta-data) from database objects
- transaction management (optional)
I think of the DBI package as the front end for interactive user at the R console, a script or package, into the other components, odbc and an ODBC driver, that make it possible to extract data from SQL server.
Role of the odbc package
The odbc package provides the DBI back-end to any odbc driver connection, including those for Microsoft SQL Server.
This enables a connection to any database with ODBC drivers available.
I think of the odbc package as the “back-end” of DBI and the “front-end” into the ODBC driver.
Role of ODBC drivers
Open Database Connectivity (ODBC) drivers are the last leg of the link between dplyr and SQL Server. They are what enable the odbc package to interface with SQL server.
I think of the SQL Server ODBC driver as the “front-end” into SQL server, and again, back to the user, script or package
Summary
- Dbplyr translates dplyr syntax into SQL code, or captures explicitly written SQL, and hands it off to DBI
- DBI uses odbc to interface with the SQL Server ODBC driver
- ODBC driver communicates with SQL server and retrieves results
User or package code -> DBI -> odbc -> SQL Server ODBC driver -> SQL server
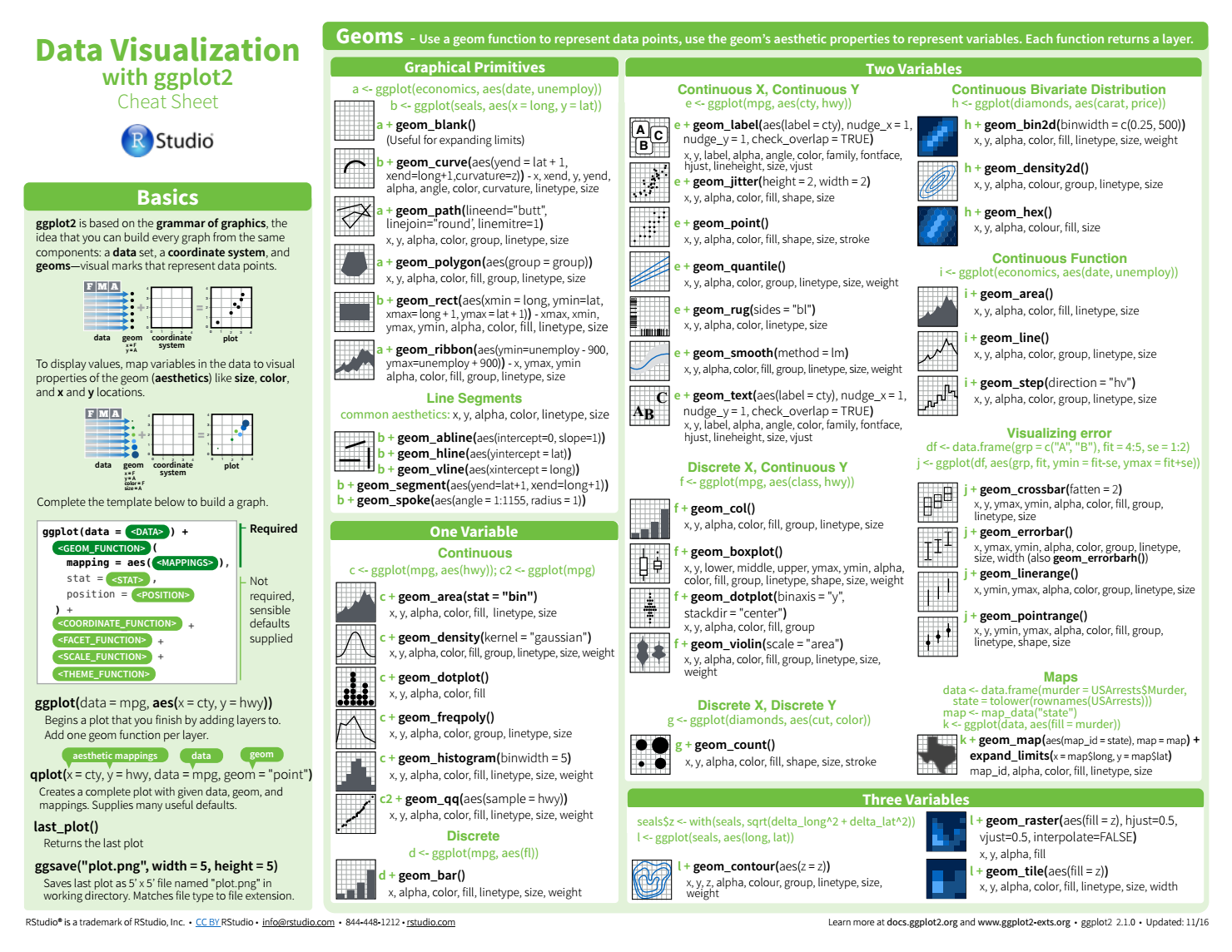
Sources and notes
I haven’t written anything new here, just focused it on the configuration I use day to day in the hopes it helps someone else.
Most was gleaned from the following and I’d recommend reviewing them for a broader perspective, and deeper insights into specific areas:
R Studio Dplyr Cheat Sheet
vignette('DBI', package = 'DBI')vignette('dbplyr', 'package = 'dbplyr)
